Multiblock supervised analyses. Should we really normalize blocks?
Multiblock supervised analyses. Should we really normalize blocks?
Abstract
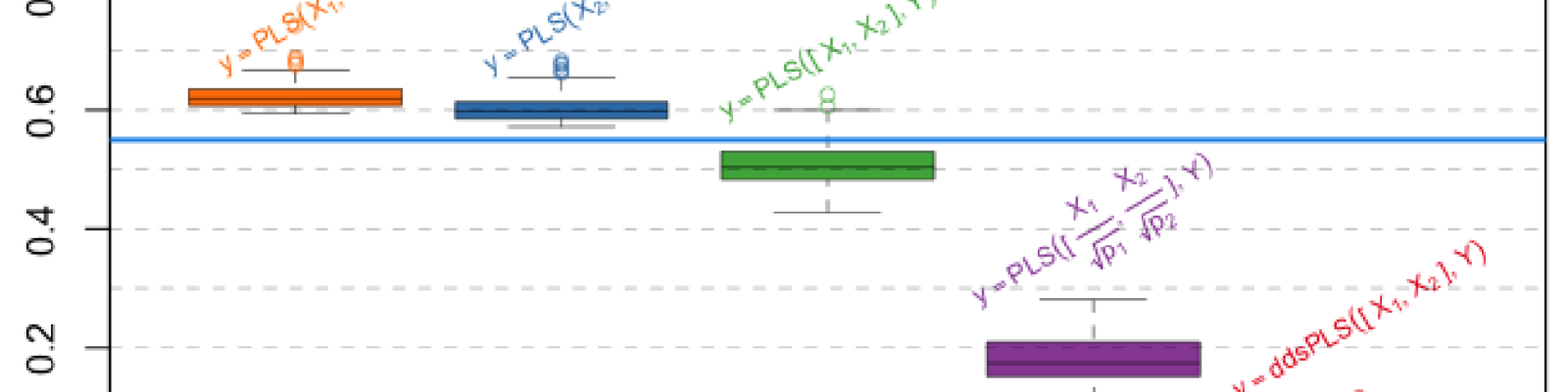
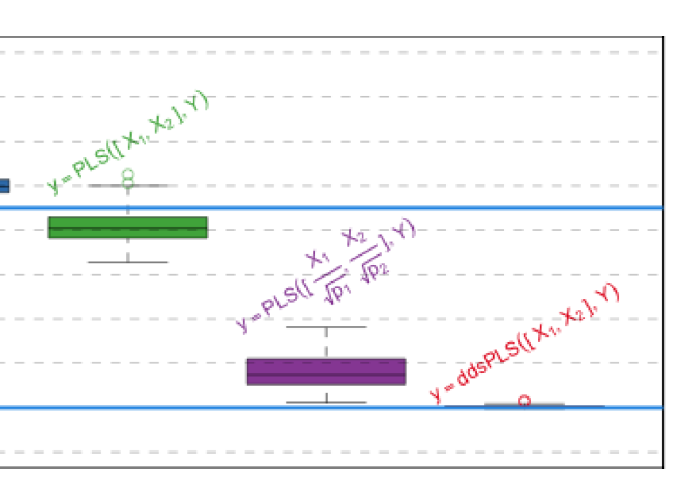
In recent years, data analysis methods have had to deal with new type of heterogeneous data sets. Multi-omics studies are perfect examples of cases where such heterogeneous data sets are obtained. While these technologies are improving in terms of accuracy, the number of variables measured simultaneously for each observation is also rising tremendously. However, these measurements are also very often carried out on very small number of observations compared to the number of variables. It has been chosen to normalize blocks by dividing each of the blocks by the largest eigenvalue, among other solutions. But no normalization can also be used. What is the best solution? We propose here to provide elements to answer this question by assessing different PLS-based methods, integrating variable selection, or not, in order to manage the large dimension of the data. We are going to show that the sparse PLS approaches provide different perspectives on how to answer this question. This study is going to be performed using simulations and real dataset applications are going to be presented.